Estimates a target parameter of interest, such as an average treatment effect (ATE), using Debiased Machine #Learning (DML).
The function dml_gate is a convenience function that adds groups to a dml object after the model is fit.
dml(
y,
d,
x,
model = c("plm", "npm"),
target = "ate",
groups = NULL,
cf.folds = 5,
cf.reps = 1,
cf.seed = NULL,
ps.trim = 0.01,
reg = "ranger",
yreg = reg,
dreg = reg,
dirty.tuning = TRUE,
save.models = FALSE,
y.class = FALSE,
d.class = FALSE,
verbose = TRUE,
warnings = FALSE
)
dml_gate(dml.fit, groups, ...)Arguments
- y
numericvector with the outcome.- d
numericvector with the treatment. If the treatment is binary, it needs to be encoded as as: zero = absence of treatment, one = presence of treatment.- x
numericvector ormatrixwith covariates. We suggest constructingxusingmodel.matrix.- model
specifies the model. Current available options are
plmfor a partially linear model, andnpmfor a fully non-parametric model.- target
specifies the target causal quantity of interest. Available options are
ate(ATE - average treatment effect),att(ATT - average treatment effect on the treated), andatu(ATU - average treatment effect on the untreated). Note that for the partially linear model with a continuous treatment the ATE also equals the average causal derivative (ACD). For the nonparametric model, these are only available for binary treatments.- groups
a
factorornumericvector indicating group membership. Groups must be a deterministic function ofx.- cf.folds
number of cross-fitting folds. Default is
5.- cf.reps
number of cross-fitting repetitions. Default is
1.- cf.seed
optional integer. A random seed for reproducibility of fold assignments.
- ps.trim
trims propensity scores lower than
ps.trimand greater than1-ps.trim, in order to obtain more stable estimates. Alternatively, a named list with elementslowerandupperspecifying the lower and upper bounds for trimming. This is only relevant for the case of a binary treatment and whenmodel = "npm".- reg
details of the machine learning method to be used for estimating the nuisance parameters (e.g, regression functions of the treatment and the outcome). Currently, this should be specified using the same arguments as
caret'strainfunction. The default is random forest usingranger. The default method is fast and usually works well for many applications.- yreg
same as
reg, but specifies arguments for the outcome regression alone. Default is the same value ofreg. Alternatively, a named list with elementsyreg0andyreg1specifying separate methods for each.- dreg
same as
reg, but specifies arguments for the treatment regression alone. Default is the same value ofreg.- dirty.tuning
should the tuning of the machine learning method happen within each cross-fit fold ("clean"), or using all the data ("dirty")? Default is dirty tuning (
dirty.tuning = TRUE). As long as the number of choices for the tuning parameters is not too big, dirty tuning is faster and should not affect the asymptotic guarantees of DML.- save.models
should the fitted models of each iterated be saved? Default is
FALSE. Note that setting this to true could end up using a lot of memory.- y.class
when
yis binary, should the outcome regression be treated as a classification problem? Default isFALSE. Note that for DML we need the class probabilities, and regression gives us that. If you change to classification, you need to make sure the method outputs class probabilities.- d.class
when
dis binary, should the outcome regression be treated as a classification problem? Default isFALSE. Note that for DML we need the class probabilities, and regression gives us that. If you change to classification, you need to make sure the method outputs class probabilities.- verbose
if
TRUE(default) prints steps of the fitting procedure.- warnings
should
caret's warnings be printed? Default isFALSE. Notecarethas many inconsistent and unnecessary warnings.- dml.fit
an object of class
dml.- ...
arguments passed to other methods.
Value
An object of class dml with the results of the DML procedure. The object is a list containing:
dataA
listwith the data used.callThe original call used to fit the model.
infoA
listwith general information and arguments of the DML fitting procedure.fitsA
listwith the predictions of each repetition.resultsA
listwith the results (influence functions and estimates) for each repetition.coefsA
listwith the estimates and standard errors for each repetition.
References
Chernozhukov, V., Cinelli, C., Newey, W., Sharma A., and Syrgkanis, V. (2026). "Long Story Short: Omitted Variable Bias in Causal Machine Learning." Review of Economics and Statistics. doi:10.1162/REST.a.1705
Examples
# loads package
library(dml.sensemakr)
## loads data
data("pension")
# set the outcome
y <- pension$net_tfa # net total financial assets
# set the treatment
d <- pension$e401 # 401K eligibility
# set the covariates (a matrix)
x <- model.matrix(~ -1 + age + inc + educ+ fsize + marr + twoearn + pira + hown, data = pension)
## compute income quartiles for group ATE.
g1 <- cut(x[,"inc"], quantile(x[,"inc"], c(0, 0.25,.5,.75,1), na.rm = TRUE),
labels = c("q1", "q2", "q3", "q4"), include.lowest = TRUE)
# run DML (nonparametric model)
## 2 folds (change as needed)
## 1 repetition (change as needed)
dml.401k <- dml(y, d, x, model = "npm", groups = g1, cf.folds = 2, cf.reps = 1)
#> Debiased Machine Learning
#>
#> Model: Nonparametric
#> Target: ate
#> Cross-Fitting: 2 folds, 1 reps
#> ML Method: outcome (yreg0:ranger, yreg1:ranger), treatment (ranger)
#> Tuning: dirty
#>
#>
#> ====================================
#> Tuning parameters using all the data
#> ====================================
#>
#> - Tuning Model for D.
#> -- Best Tune:
#> mtry min.node.size splitrule
#> 1 2 5 variance
#>
#> - Tuning Model for Y (non-parametric).
#> -- Best Tune:
#> mtry min.node.size splitrule
#> 1 2 5 variance
#> mtry min.node.size splitrule
#> 1 2 5 variance
#>
#>
#> ======================================
#> Repeating 2-fold cross-fitting 1 times
#> ======================================
#>
#> -- Rep 1 -- Folds: 1 2
#>
# summary of results with median method (default)
summary(dml.401k, combine.method = "median")
#>
#> Debiased Machine Learning
#>
#> Model: Nonparametric
#> Cross-Fitting: 2 folds, 1 reps
#> ML Method: outcome (yreg0:ranger, yreg1:ranger, R2 = 26.034%), treatment (ranger, R2 = 11.251%)
#> Tuning: dirty
#>
#> Average Treatment Effect:
#>
#> Estimate Std. Error t value P(>|t|)
#> ate.all 7763 1145 6.781 1.2e-11 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Group Average Treatment Effect:
#>
#> Estimate Std. Error t value P(>|t|)
#> gate.q1 4772 840 5.681 1.34e-08 ***
#> gate.q2 3093 1217 2.541 0.011048 *
#> gate.q3 6330 1760 3.596 0.000323 ***
#> gate.q4 16854 3955 4.262 2.03e-05 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Note: DML estimates combined using the median method.
#>
#> Verbal interpretation of DML procedure:
#>
#> -- Average treatment effects were estimated using DML with 2-fold cross-fitting. In order to reduce the variance that stems from sample splitting, we repeated the procedure 1 times. Estimates are combined using the median as the final estimate, incorporating variation across experiments into the standard error as described in Chernozhukov et al. (2018). The outcome regression uses from the R package ; the treatment regression uses Random Forest from the R package ranger.
# coef median method (default)
coef(dml.401k, combine.method = "median")
#> ate.all gate.q1 gate.q2 gate.q3 gate.q4
#> 7762.915 4772.061 3093.195 6330.041 16854.429
# se median method (default)
se(dml.401k, combine.method = "median")
#> ate.all gate.q1 gate.q2 gate.q3 gate.q4
#> 1144.8702 839.9974 1217.2293 1760.0668 3954.6826
# confint median method
confint(dml.401k, combine.method = "median")
#> 2.5 % 97.5 %
#> ate.all 5519.0104 10006.819
#> gate.q1 3125.6963 6418.426
#> gate.q2 707.4699 5478.921
#> gate.q3 2880.3735 9779.708
#> gate.q4 9103.3935 24605.465
## Sensitivity Analysis
### Robustness Values
robustness_value(dml.401k, alpha = 0.05)
#> ate.all gate.q1 gate.q2 gate.q3 gate.q4
#> 0.04377913 0.07882806 0.01530738 0.03302966 0.05237003
### Confidence Bounds
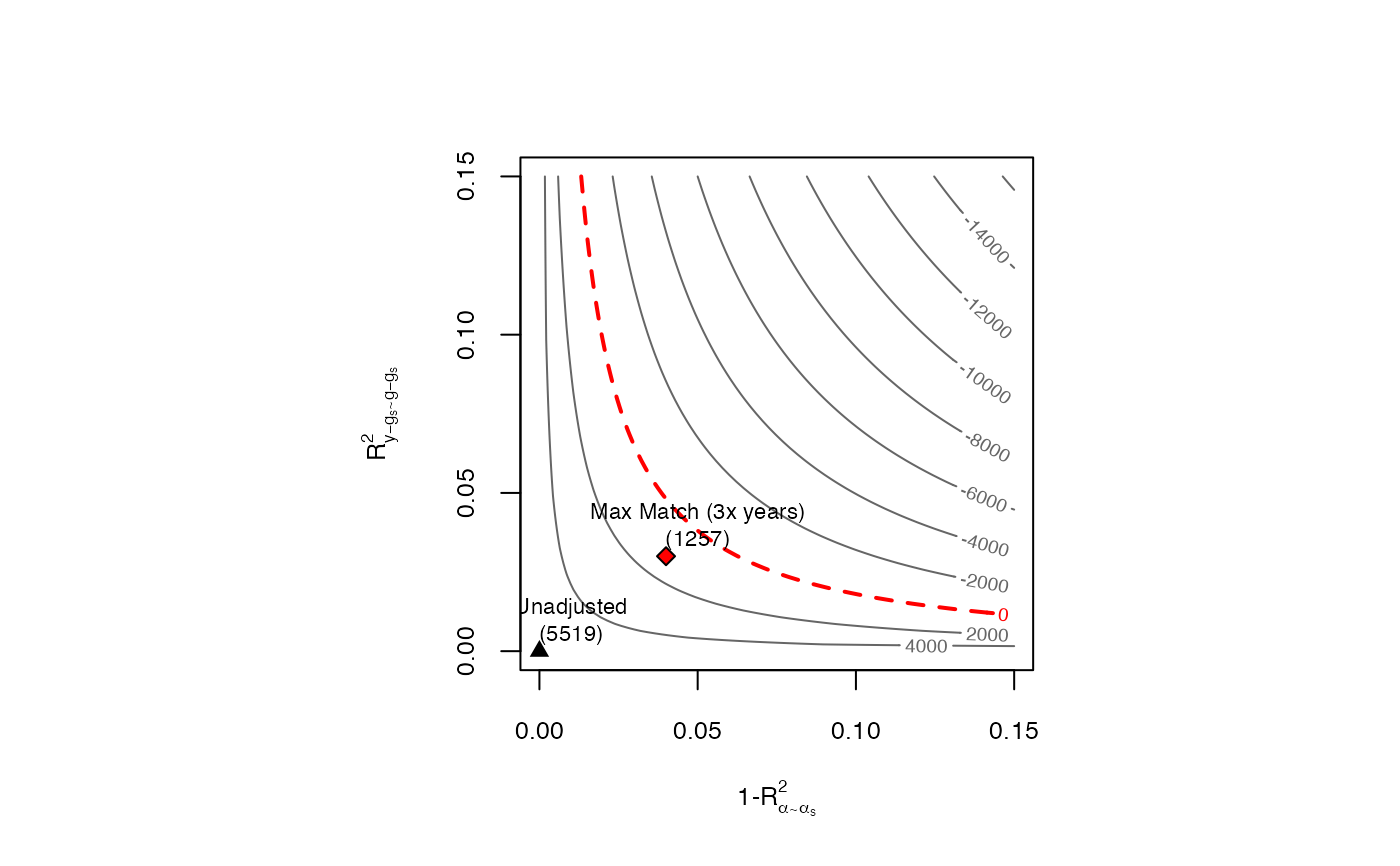
confidence_bounds(dml.401k, cf.y = 0.03, cf.d = 0.04, level = 0.95)
#> lwr upr
#> ate.all 1256.9657 14264.6146
#> gate.q1 1951.1698 7711.1079
#> gate.q2 -1451.3430 6982.7950
#> gate.q3 -187.0972 12637.9706
#> gate.q4 3587.5139 30200.0130
#>
#> Confidence level: point = 95%; region = 90%.
#> Sensitivity parameters: cf.y = 0.03; cf.d = 0.04; rho2 = 1.
### Contour Plots
ovb_contour_plot(dml.401k, cf.y = 0.03, cf.d = 0.04,
bound.label = "Max Match (3x years)")