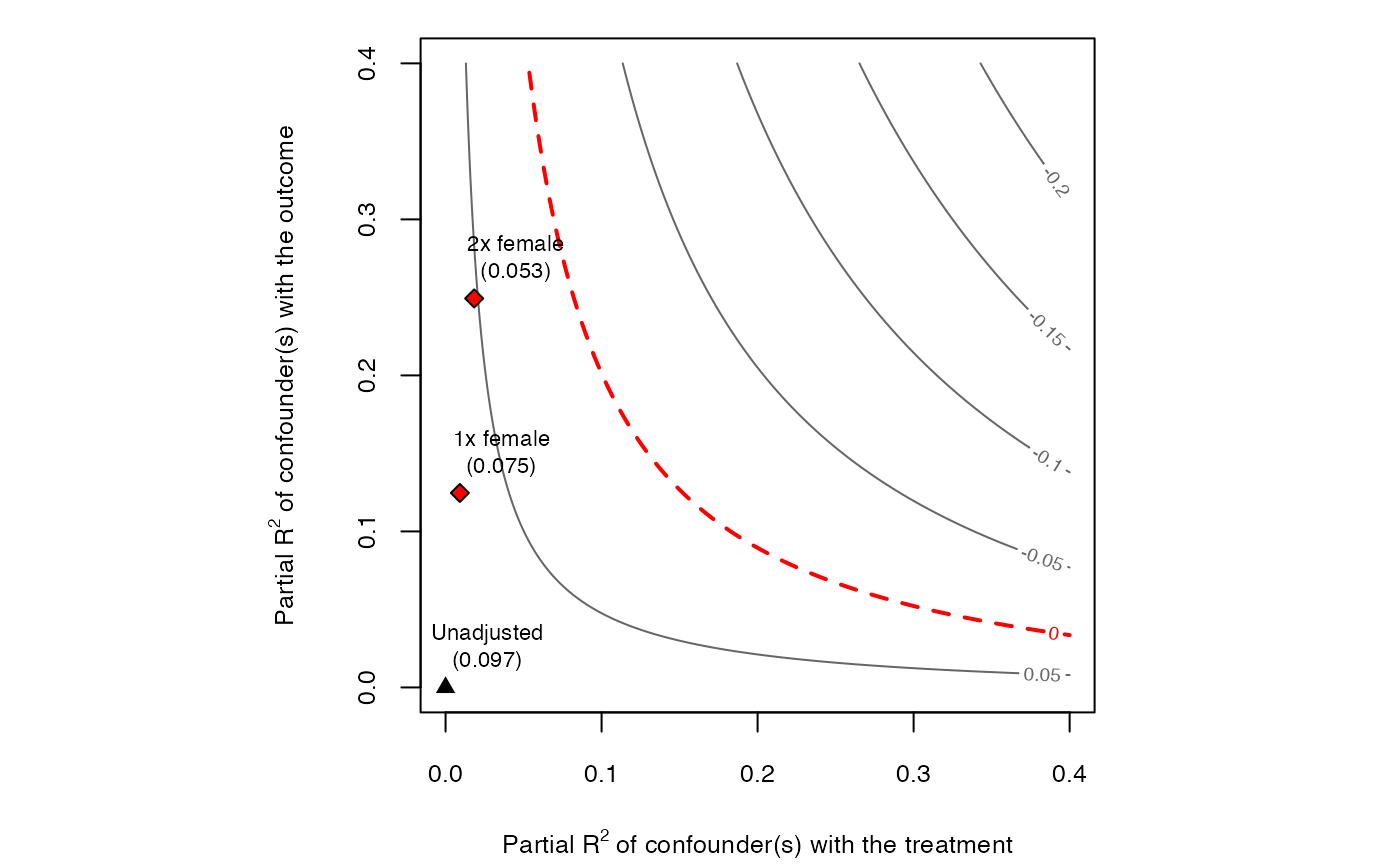
Contour plots of omitted variable bias for sensitivity analysis. The main inputs are an lm model, the treatment variable
and the covariates used for benchmarking the strength of unobserved confounding.
The horizontal axis of the plot shows hypothetical values of the partial R2 of the unobserved confounder(s) with the treatment.
The vertical axis shows hypothetical values of the partial R2 of the unobserved confounder(s) with the outcome.
The contour levels represent the adjusted estimates (or t-values) of the treatment effect.
The reference points are the bounds on the partial R2 of the unobserved confounder if it were k times ``as strong'' as the observed covariate used for benchmarking (see arguments kd and ky).
The dotted red line show the chosen critical threshold (for instance, zero): confounders with such strength (or stronger) are sufficient to invalidate the research conclusions.
All results are exact for single confounders and conservative for multiple/nonlinear confounders.
See Cinelli and Hazlett (2020) for details.
ovb_contour_plot(model, ...)
# S3 method for lm
ovb_contour_plot(
model,
treatment,
benchmark_covariates = NULL,
kd = 1,
ky = kd,
r2dz.x = NULL,
r2yz.dx = r2dz.x,
bound_label = "manual",
sensitivity.of = c("estimate", "t-value"),
reduce = TRUE,
estimate.threshold = 0,
t.threshold = 2,
nlevels = 10,
col.contour = "grey40",
col.thr.line = "red",
label.text = TRUE,
cex.label.text = 0.7,
round = 3,
...
)
# S3 method for fixest
ovb_contour_plot(
model,
treatment,
benchmark_covariates = NULL,
kd = 1,
ky = kd,
r2dz.x = NULL,
r2yz.dx = r2dz.x,
bound_label = "manual",
sensitivity.of = c("estimate", "t-value"),
reduce = TRUE,
estimate.threshold = 0,
t.threshold = 2,
nlevels = 10,
col.contour = "grey40",
col.thr.line = "red",
label.text = TRUE,
cex.label.text = 0.7,
round = 3,
...
)
# S3 method for formula
ovb_contour_plot(
formula,
method = c("lm", "feols"),
vcov = "iid",
data,
treatment,
benchmark_covariates = NULL,
kd = 1,
ky = kd,
r2dz.x = NULL,
r2yz.dx = r2dz.x,
bound_label = NULL,
sensitivity.of = c("estimate", "t-value"),
reduce = TRUE,
estimate.threshold = 0,
t.threshold = 2,
nlevels = 10,
col.contour = "grey40",
col.thr.line = "red",
label.text = TRUE,
cex.label.text = 0.7,
round = 3,
...
)
# S3 method for numeric
ovb_contour_plot(
estimate,
se,
dof,
r2dz.x = NULL,
r2yz.dx = r2dz.x,
bound_label = rep("manual", length(r2dz.x)),
sensitivity.of = c("estimate", "t-value"),
reduce = TRUE,
estimate.threshold = 0,
t.threshold = 2,
show.unadjusted = TRUE,
lim = NULL,
lim.y = NULL,
nlevels = 10,
col.contour = "black",
col.thr.line = "red",
label.text = TRUE,
cex.label.text = 0.7,
label.bump.x = NULL,
label.bump.y = NULL,
xlab = NULL,
ylab = NULL,
cex.lab = 0.8,
cex.axis = 0.8,
cex.main = 1,
asp = lim/lim.y,
list.par = list(mar = c(4, 4, 1, 1), pty = "s"),
round = 3,
...
)Arguments
- model
An
fixestobject with the outcome regression.- ...
arguments passed to other methods. First argument should either be an
lmmodel with the outcome regression, aformuladescribing the model along with thedata.framecontaining the variables of the model, or a numeric vector with the coefficient estimate.- treatment
A character vector with the name of the treatment variable of the model.
- benchmark_covariates
The user has two options: (i) character vector of the names of covariates that will be used to bound the plausible strength of the unobserved confounders. Each variable will be considered separately; (ii) a named list with character vector names of covariates that will be used, as a group, to bound the plausible strength of the unobserved confounders. The names of the list will be used for the benchmark labels. Note: for factor variables with more than two levels, you need to provide the name of each level as encoded in the
fixestmodel (the columns ofmodel.matrix).- kd
numeric vector. Parameterizes how many times stronger the confounder is related to the treatment in comparison to the observed benchmark covariate. Default value is
1(confounder is as strong as benchmark covariate).- ky
numeric vector. Parameterizes how many times stronger the confounder is related to the outcome in comparison to the observed benchmark covariate. Default value is the same as
kd.- r2dz.x
Hypothetical partial R2 of unobserved confounder Z with treatment D, given covariates X.
- r2yz.dx
Hypothetical partial R2 of unobserved confounder Z with outcome Y, given covariates X and treatment D.
- bound_label
label to bounds provided manually in
r2dz.xandr2yz.dx.- sensitivity.of
should the contour plot show adjusted estimates (
"estimate") or adjusted t-values ("t-value")?- reduce
Should the bias adjustment reduce or increase the absolute value of the estimated coefficient? Default is
TRUE.- estimate.threshold
critical threshold for the point estimate.
- t.threshold
critical threshold for the t-value.
- nlevels
number of levels for the contour plot.
- col.contour
color of contour lines.
- col.thr.line
color of threshold contour line.
- label.text
should label texts be plotted? Default is
TRUE.- cex.label.text
size of the label text.
- round
number of digits to show in contours and bound values
- formula
an object of the class
formula: a symbolic description of the model to be fitted.- method
the default is
lm. This argument can be changed to estimate the model usingfeols. In this case the formula needs to be written so it can be estimated withfeolsand the package needs to be installed.- vcov
the variance/covariance used in the estimation when using
feols. Seevcov.fixestfor more details. Defaults to "iid".- data
data needed only when you pass a formula as first parameter. An object of the class
data.framecontaining the variables used in the analysis.- estimate
Coefficient estimate.
- se
Standard error of the coefficient estimate.
- dof
Residual degrees of freedom of the regression.
- show.unadjusted
should the unadjusted estimates be shown? Default is `TRUE`.
- lim
sets limit for x-axis. If `NULL`, limits are computed automatically.
- lim.y
sets limit for y-axis. If `NULL`, limits are computed automatically.
- label.bump.x
bump on the x coordinate of label text.
- label.bump.y
bump on the y coordinate of label text.
- xlab
label of x axis. If `NULL`, default label is used.
- ylab
label of y axis. If `NULL`, default label is used.
- cex.lab
The magnification to be used for x and y labels relative to the current setting of cex.
- cex.axis
The magnification to be used for axis annotation relative to the current setting of cex.
- cex.main
The magnification to be used for main titles relative to the current setting of cex.
- asp
the y/x aspect ratio. Default is 1.
- list.par
arguments to be passed to
par. It needs to be a named list.
Value
The function returns invisibly the data used for the contour plot (contour grid and bounds).
References
Cinelli, C. and Hazlett, C. (2020), "Making Sense of Sensitivity: Extending Omitted Variable Bias." Journal of the Royal Statistical Society, Series B (Statistical Methodology).
Examples
# runs regression model
model <- lm(peacefactor ~ directlyharmed + age + farmer_dar + herder_dar +
pastvoted + hhsize_darfur + female + village,
data = darfur)
# contour plot
ovb_contour_plot(model, treatment = "directlyharmed",
benchmark_covariates = "female",
kd = 1:2)